import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
Package Version
---------- ---------
python 3.8.8
matplotlib 3.4.3
seaborn 0.11.1
Plot types
Scatter plot
Basic
def plot_something(srs: pd.Series) -> None:
fig, ax = plt.subplots(figsize=(4, 4), dpi=100)
ax.scatter(srs.index, srs.values, label='', zorder=2)
ax.set_title('')
ax.set_ylabel('')
ax.set_xlabel('')
ax.grid()
plt.savefig('chart.png')
srs = pd.Series(np.random.randint(20, size=10))
plot_something(srs)
Labelled scatter plot near intercept
Here’s an example of a simple scatter plot using labeled data points centered around the intercept. I initially wrote this as a base for a function which visualizes how the k-means algorithm works step-by-step. A couple things to note:
- We set higher
zorderfor the elements we want to appear “on top” of others - The core
ax.scatterfunction takes an array of data points, butax.annotatedoes not, so we need to loop.
data = np.asarray([
[2, 2],
[-1, 1],
[3, 1],
[0, -1],
[-2, -2]
])
def scatter_plot_near_intercept(data):
fig, ax = plt.subplots(figsize=(5, 5), dpi=100)
# Format grid
ax.grid(zorder=1)
ax.axvline(0, c='black', zorder=2)
ax.axhline(0, c='black', zorder=2)
ax.set_xlim(-4, 4)
ax.set_ylim(-4, 4)
# Plot data points
ax.scatter(*data.T, s=256, c='yellow', edgecolors='black', zorder=3)
for i, (x, y) in enumerate(data):
ax.annotate(i, (x, y), size=10, ha='center', va='center')
scatter_plot_near_intercept(data)
Plotting functions
Plot a pdf
x = np.arange(-4, 4, 0.1)
def plot_pdf(x, y, title=None) -> None:
fig, ax = plt.subplots(figsize=(4, 4), dpi=100)
ax.plot(x, y)
ax.set_title(title)
ax.set_ylabel('')
ax.set_xlabel('')
ax.grid()
ax.axvline(0, c='black', linewidth=1, zorder=2)
ax.axhline(0, c='black', linewidth=1, zorder=2)
plt.savefig('chart.png')
plot_pdf(x, x*np.exp(-x**2/2), r'Plot of $z e^{-\frac{z^2}{2}}$')
Plot multiple pdfs
from scipy.stats import norm
def plot_pdfs(x, pdfs: dict) -> None:
fig, ax = plt.subplots(figsize=(4, 4), dpi=100)
for label, pdf in pdfs.items():
ax.plot(x, pdf, label=label)
ax.set_title('')
ax.set_ylabel('')
ax.set_xlabel('')
ax.legend(loc='best')
ax.grid()
ax.axvline(0, c='black', linewidth=1, zorder=2)
ax.axhline(0, c='black', linewidth=1, zorder=2)
plt.savefig('chart.png')
x = np.arange(-3, 3, 0.1)
log_lk = lambda z: -(1/2)*np.log(2*np.pi) - z**2/2
dldz = lambda z: -z
dldz_sqr = lambda z: z**2
pdfs = {
r'$f(x|\theta)$': norm.pdf(x),
r'$\ell(x|θ)$': log_lk(x),
r'$\frac{d \ell}{dx}(x|θ)$': dldz(x),
r'$[\frac{d \ell}{dx}(x|θ)]^2$': dldz_sqr(x)
}
plot_pdfs(x, pdfs)
Discontinuous function
def plot_discontinuous_function(x, y, discontinuities=None, title=None) -> None:
fig, ax = plt.subplots(figsize=(4, 4), dpi=100)
for dc in discontinuities:
x_seg = x[x<dc]
y_seg = np.compress(x<dc, y)
ax.plot(x_seg, y_seg, color='blue', zorder=3)
ax.scatter(x_seg[-1], y_seg[-1], marker='o', zorder=4, facecolor='white', edgecolor='blue')
# plot data between final discontinuity and end of series
x_seg = x[x>dc]
y_seg = np.compress(x>dc, y)
ax.plot(x_seg, y_seg, color='blue', zorder=3)
ax.scatter(x_seg[0], y_seg[0], marker='o', zorder=4, facecolor='white', edgecolor='blue')
ax.set_title(title)
ax.set_ylabel('')
ax.set_xlabel('')
ax.grid()
ax.axvline(0, c='black', linewidth=1, zorder=2)
ax.axhline(0, c='black', linewidth=1, zorder=2)
plt.savefig('chart.png')
x = np.arange(-2, 2, 0.01)
y = 1/x - 1/np.abs(x)
plot_discontinuous_function(x, y, [0], r'')
Time series
def plot_time_series(srs: pd.Series) -> None:
""" Plot a simple time series (with moving average) using matplotlib """
fig, ax = plt.subplots(figsize=(10, 4))
ax.grid(True)
fig.set_facecolor('white')
ax.plot(srs, color='tab:grey', alpha=0.3)
ma = srs.rolling(window=7, min_periods=1).mean()
ax.plot(ma, color='tab:blue')
idx = pd.date_range('2021-01-01', '2021-07-01')
data = np.arange(50, 50+len(idx)) + np.random.normal(0, 10, size=len(idx))
srs = pd.Series(data, index=idx)
plot_time_series(srs)
Formatting
from scipy.stats import norm
def basic_plot():
fig, ax = plt.subplots(figsize=(4, 4), dpi=100)
x = np.arange(-3, 3, 0.1)
y = norm.pdf(x)
ax.plot(x, y)
return fig, ax
basic_plot()
(<Figure size 400x400 with 1 Axes>, <AxesSubplot:>)
Legend
Axis formatting
Percent
https://matplotlib.org/stable/api/ticker_api.html#matplotlib.ticker.PercentFormatter
fig, ax = basic_plot()
import matplotlib.ticker as mtick
ax.yaxis.set_major_formatter(mtick.PercentFormatter(xmax=1, decimals=0))
from matplotlib.ticker import FuncFormatter
def currency(x, pos):
'The two args are the value and tick position'
if x >= 1000000:
return '${:1.1f}M'.format(x*1e-6)
return '${:1.0f}K'.format(x*1e-3)
formatter = FuncFormatter(currency)
ax.xaxis.set_major_formatter(formatter)
Dual Y axis
def add_relabelled_y_axis_to_right_side(ax, new_labels: list):
""" Add a set of labels to the RHS of a plot, to aid in interpretability. """
label_objs = list(ax.get_yticklabels())
for label, orig in zip(label_objs, new_labels):
label._text = orig
# create a new y-axis on RHS
ax2 = ax.twinx()
# Copy tick locations
_ = ax2.set_yticks(ax.get_yticks())
# Copy tick labels
_ = ax2.set_yticklabels(label_objs)
# Copy axis limits
_ = ax2.set_ylim(ax.get_ylim())
# Resize RHS ticks to match LHS
if 'labelsize' in ax.yaxis._major_tick_kw:
label_size = ax.yaxis._major_tick_kw['labelsize']
for tick in ax2.yaxis.majorTicks:
tick.label2.set_fontsize(label_size)
return ax
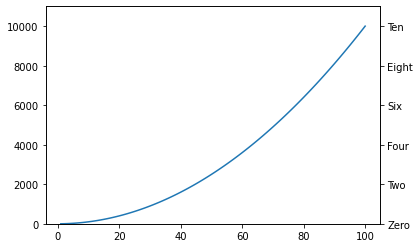
fig, ax = plt.subplots()
x = np.arange(1, 101)
ax.plot(x, x**2)
ax.set_ylim(0, 11000)
add_relabelled_y_axis_to_right_side(ax, ['Zero', 'Two', 'Four', 'Six', 'Eight', 'Ten']);
def dual_axis_line_chart(
x: pd.Series,
y1: pd.Series,
y2: pd.Series,
title: str = None,
ylabel: str = None,
xlabel: str = None,
) -> None:
""" """
fig, ax1 = plt.subplots(figsize=(9, 6))
ax1.plot(x, y1)
ax2 = ax1.twinx()
ax2.plot(x, y2)
if title:
ax1.set_title(title)
if ylabel:
ax1.set_ylabel(ylabel)
if xlabel:
ax1.set_xlabel(xlabel)
Multiple plots
Layouts
Multiple rows and columns
# larger grid
fig, axes = plt.subplots(
figsize=(6, 6),
nrows=2,
ncols=2,
sharex=True,
sharey=True,
gridspec_kw={'wspace': 0.1},
dpi=100
)
axes = axes.flat
Unqual sized
fig, (ax1, ax2) = plt.subplots(
nrows=2,
sharex=True,
gridspec_kw={'height_ratios': [3, 1]},
figsize=(18, 9),
dpi=100
)

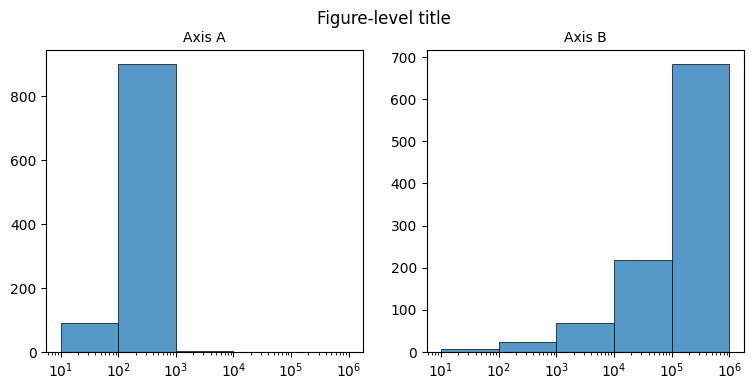
Boilerplate
# Set up figure
fig, axes = plt.subplots(
figsize=(9, 4),
nrows=1,
ncols=2,
gridspec_kw={'top': 0.88},
dpi=100
)
ax1, ax2 = list(axes.flat)
plt.style.use('default')
_ = fig.suptitle('Figure-level title')
# Generate data
x = np.arange(1, 1001)
srs_A = pd.Series(x, index=x)
srs_B = pd.Series(x**2, index=x)
# Axis A
_ = sns.histplot(
srs_A,
log_scale=True,
linewidth=0.5,
bins=[1, 2, 3, 4, 5, 6],
ax=ax1
)
_ = ax1.set_title(f'Axis A', size=10)
_ = ax1.set_xlabel('')
_ = ax1.set_ylabel('')
# Axis B
_ = sns.histplot(
srs_B,
log_scale=True,
linewidth=0.5,
bins=[1, 2, 3, 4, 5, 6],
ax=ax2
)
_ = ax2.set_title(f'Axis B', size=10)
_ = ax2.set_xlabel('')
_ = ax2.set_ylabel('')
Human-readable numbers
Proof of concept
from math import log, floor
def human_format(number):
""" Source: https://stackoverflow.com/questions/579310/formatting-long-numbers-as-strings-in-python/45846841 """
units = ['', 'K', 'M', 'B']
k = 1000
try:
magnitude = int(floor(log(number, k)))
return f'{number/k**magnitude:.0f}{units[magnitude]}'
except ValueError:
return '0'
for x in range(1, 12):
print(f'{10**x:12} {human_format(10**x)}')
10 10
100 100
1000 1K
10000 10K
100000 100K
1000000 1M
10000000 10M
100000000 100M
1000000000 1B
10000000000 10B
100000000000 100B
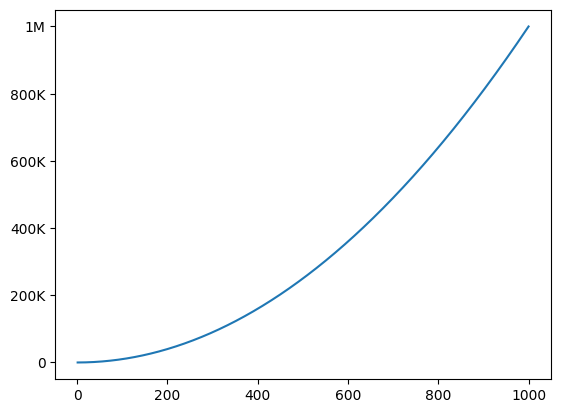
In practice
from math import floor, log
from matplotlib.ticker import FuncFormatter
# generate dummy plot
fig, ax = plt.subplots()
x = np.arange(1, 1001)
ax.plot(x, x**2)
ax.ticklabel_format(style='plain') # disable scientific notation
def human_format(number, pos):
units = ['', 'K', 'M', 'B']
k = 1000
try:
magnitude = int(floor(log(number, k)))
return f'{number/k**magnitude:.0f}{units[magnitude]}'
except ValueError:
return '0'
# apply formatter
ax.yaxis.set_major_formatter(FuncFormatter(human_format))
Unsorted
fig, ax = plt.subplots()
x = np.arange(1, 101)
ax.plot(x, x**2)
ax.set_ylim(0, 11000)
ax.axhline(2000, linestyle='--', color='purple')
<matplotlib.lines.Line2D at 0x7f7f018f9700>
Other
Auto-tilt x-axis labels
fig.autofmt_xdate()
Change interval of dates on x-axis
import matplotlib.dates as mdates
ax.xaxis.set_major_locator(mdates.MonthLocator(interval=1))
ax.xaxis.set_major_formatter(mdates.DateFormatter('%Y-%m-%d'))
Further reading
Real Python: Python Plotting With Matplotlib – Great introductory resource, with an explanation of the two different APIs, which is a key source of confusion for new users.
Python Data Science Handbook: Visualization with Matplotlib – Collection of 10+ notebooks with details on how to achieve specific tweaks.