import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.preprocessing import PowerTransformer
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LinearRegression, Ridge, RidgeCV
from sklearn.model_selection import GridSearchCV, KFold
Package Version
------------ ---------
python 3.9.12
numpy 1.21.5
pandas 1.4.2
matplotlib 3.5.1
scikit-learn 1.0.2
General
Train–Test split
from sklearn.model_selection import train_test_split
from sklearn.datasets import make_regression
X, y = make_regression(n_features=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.20)
Feature engineering
Apply scikit-learn preprocessing with keeping index and columns of DataFrame
Scikit-learn includes a bunch of useful feature transformation functions such as PolynomialFeatures and OneHotEncoder. They are easy to use as part of a model pipeline, but their intermediate outputs (numpy matrices) can be difficult to interpret. I find havng these intermediate outputs back in a pandas DataFrame with the original index and column names is helpful when troubleshooting or communicating the results of preprocessing.
from sklearn.preprocessing import PolynomialFeatures, FunctionTransformer
df = pd.DataFrame({'category': ['a', 'b', 'c'], 'numeric': np.random.random(size=3)})
display(df)
def pandas_polynomial_features(X):
""" Wrapper for calling PolynomialFeatures while preserving
index and column names from input DataFrame. """
poly_tx = PolynomialFeatures(degree=2, include_bias=False).fit(X)
poly_cols = poly_tx.get_feature_names_out(input_features=X.columns)
return pd.DataFrame(poly_tx.transform(X), index=X.index, columns=poly_cols)
PandasPolynomialFeatures = FunctionTransformer(pandas_polynomial_features, validate=False)
numeric_cols = df.select_dtypes(np.number).dtypes.index.values.tolist()
PandasPolynomialFeatures.fit_transform(df[numeric_cols])
| category | numeric | |
|---|---|---|
| 0 | a | 0.753072 |
| 1 | b | 0.391670 |
| 2 | c | 0.245304 |
| numeric | numeric^2 | |
|---|---|---|
| 0 | 0.753072 | 0.567117 |
| 1 | 0.391670 | 0.153405 |
| 2 | 0.245304 | 0.060174 |
Getting readable metadata from OneHotEncoder
Fitted OneHotEncoder objects do have a categories_ attribute, but it only includes the “child” value without reference to the “parent” categorical variable. If you are working with categoricals whose values overlap (e.g. departure and arrival city of a trip) this can become confusing. I prefer the column names to include the initials of the initial column separated by _is_, to aid in interpretability during later phases.
df = pd.DataFrame({'home_city': np.random.choice(['toronto', 'paris', 'london'], size=5),
'fav_city': np.random.choice(['toronto', 'paris', 'london'], size=5)}, dtype='category')
display(df)
| home_city | fav_city | |
|---|---|---|
| 0 | paris | paris |
| 1 | toronto | toronto |
| 2 | paris | london |
| 3 | london | toronto |
| 4 | toronto | paris |
from sklearn.preprocessing import OneHotEncoder
def pandas_one_hot_encoder(X):
""" Wrapper for calling PolynomialFeatures while preserving index and column names from input DataFrame. """
fit_obj = OneHotEncoder().fit(X)
nested_output_col_names = fit_obj.categories_
flat_output_col_names = []
for cat_col, corresp_enc_cols in zip(X.columns, nested_output_col_names):
stem = ''.join([x[0] for x in cat_col.split('_')]) + '_is_'
flat_output_col_names.extend(stem + corresp_enc_cols)
return pd.DataFrame(fit_obj.transform(X).toarray(),
index=X.index,
columns=flat_output_col_names)
PandasOneHotEncoder = FunctionTransformer(pandas_one_hot_encoder, validate=False)
PandasOneHotEncoder.fit_transform(df).head()
| hc_is_london | hc_is_paris | hc_is_toronto | fc_is_london | fc_is_paris | fc_is_toronto | |
|---|---|---|---|---|---|---|
| 0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 1 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 |
| 2 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 3 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 4 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 |
Using ColumnTransformer from inside a pipeline
Scikit-learn has recently introduced ColumnTransformer , which is a very useful tool for performing preprocessing on a dataset with heterogeneous dtypes. Previously this was cumbersome without using something like sklearn-pandas. It is not yet perfectly integrated with the rest of the sklearn API though. Recently I was trying to use ColumnTransformer as part of a Pipeline object, but encountered the following error:
ValueError: Specifying the columns using strings is only supported for pandas DataFrames
The transformers parameter of ColumnTransformer takes a list of three-element tuples, the third of which points towards which columns should be passed into each respective step. When you pass a pandas DataFrame directly to the transformer via its .fit() method, you can use a list of column names as this third element in each tuple. But if you are using ColumnTransformer as part of a pipeline, these have to be numeric indices.
df = pd.DataFrame({'home_city': np.random.choice(['toronto', 'paris', 'london'], size=5),
'fav_city': np.random.choice(['toronto', 'paris', 'london'], size=5)}, dtype='category')
df['age'] = np.random.randint(80, size=5)
display(df)
numeric_cols = df.dtypes.apply(lambda x: x.kind in 'bifc').reset_index(drop=True).loc[lambda x: x == True].index
cat_cols = (df.dtypes == 'category').reset_index(drop=True).loc[lambda x: x == True].index
col_tx = ColumnTransformer(transformers=[
('num', PowerTransformer(), numeric_cols),
('cat', OneHotEncoder(drop='first', handle_unknown='error'), cat_cols)
])
pipe = Pipeline([
('col_tx', col_tx),
('model', LinearRegression())
])
pipe.fit(df, [0, 1, 0, 1, 0]);
| home_city | fav_city | age | |
|---|---|---|---|
| 0 | london | toronto | 79 |
| 1 | london | paris | 39 |
| 2 | toronto | paris | 11 |
| 3 | toronto | london | 3 |
| 4 | paris | paris | 3 |
Parameter search on pre-processing steps
It is straightforward to use GridSearchCV on final model parameters, but it is also possible to use it for parameters of “upstream” data transformations, if you compose them together with the final model into a Pipeline object. Then you can use use the syntax <estimator>__<param> separated by a double underscore in your param_dict keys to specify which parameter belongs to which step of the pipeline. Read more at the official scikit-learn docs.
from sklearn.datasets import make_regression
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
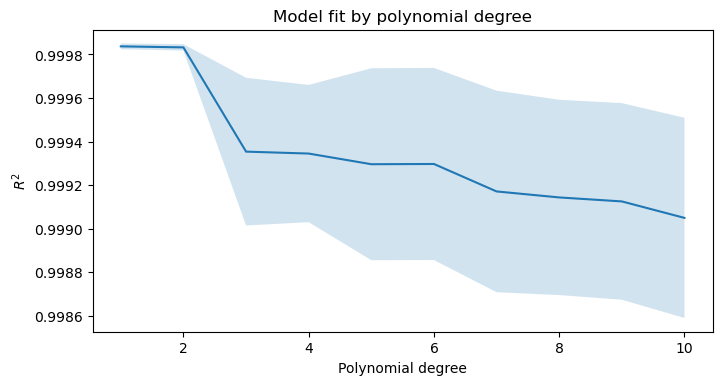
def find_best_degrees(X, y, degrees=range(1, 11)):
""" Example of grid search on parameters of an upstream transformation step in a pipeline. """
pipe = Pipeline([
('poly_tx', PolynomialFeatures(include_bias=False)),
('model', make_pipeline(StandardScaler(with_mean=False), Ridge()))
])
param_grid = {'poly_tx__degree': degrees}
search = GridSearchCV(pipe, param_grid, cv=KFold(n_splits=5, shuffle=True)).fit(X, y)
avg_scores = search.cv_results_['mean_test_score']
std_scores = search.cv_results_['std_test_score']
fig, ax = plt.subplots(figsize=(8, 4), dpi=100)
ax.plot(degrees, avg_scores, label='$R^2$')
ax.fill_between(degrees, avg_scores-std_scores, avg_scores+std_scores, alpha=0.2)
ax.set_title('Model fit by polynomial degree')
ax.set_ylabel('$R^2$')
ax.set_xlabel('Polynomial degree')
X, y = make_regression(n_features=1)
find_best_degrees(X, y)
comments powered by Disqus